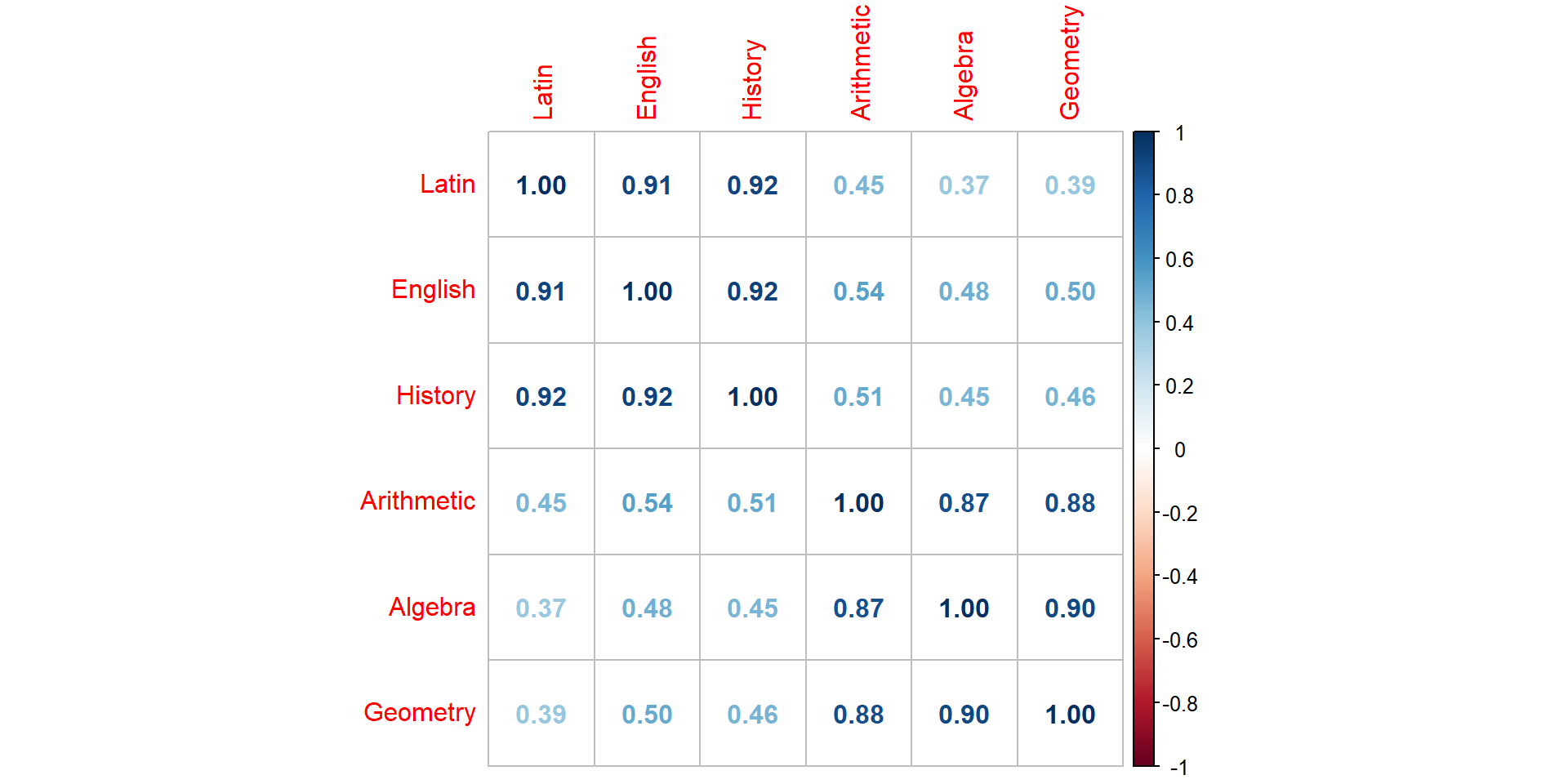
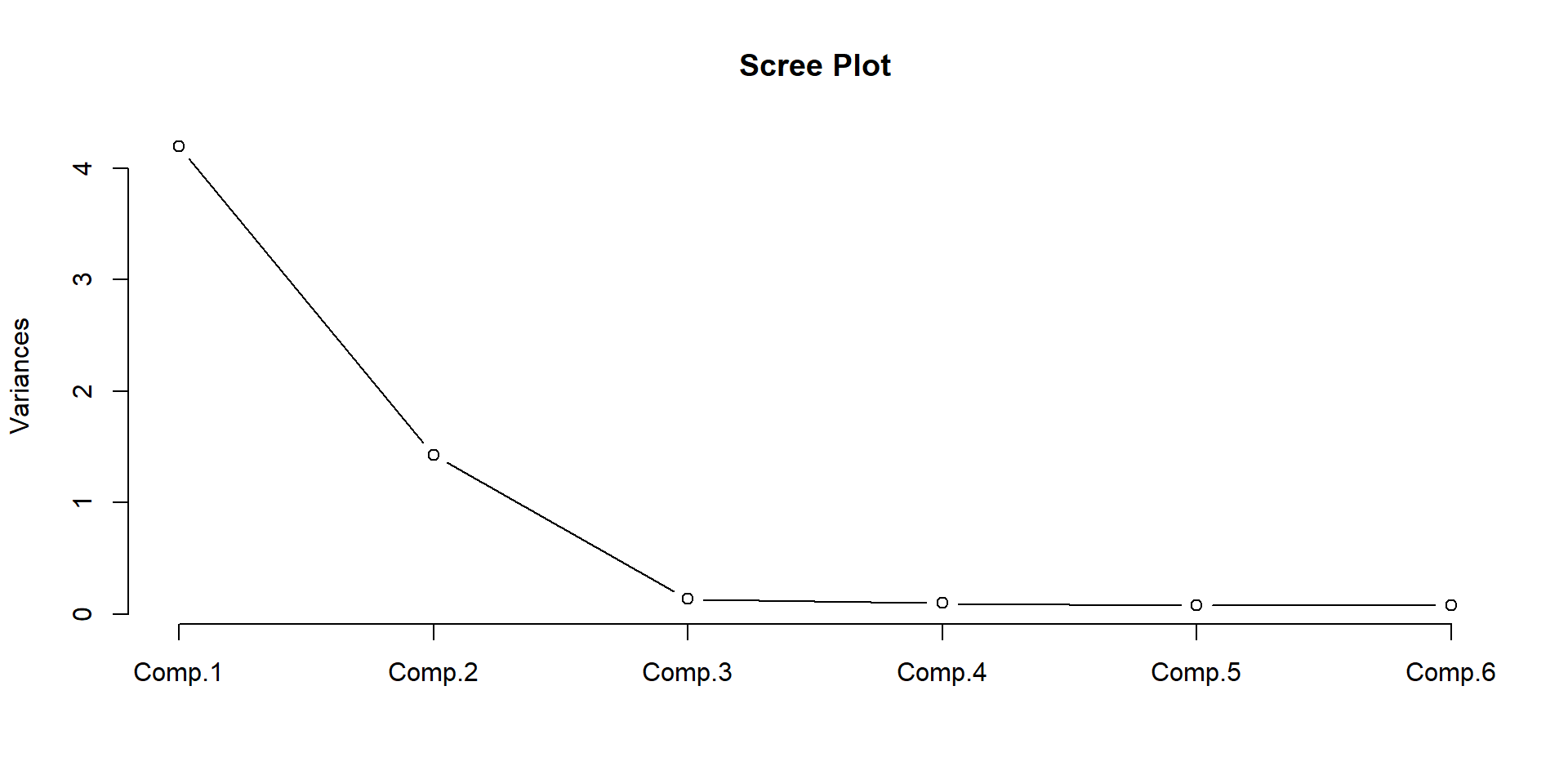
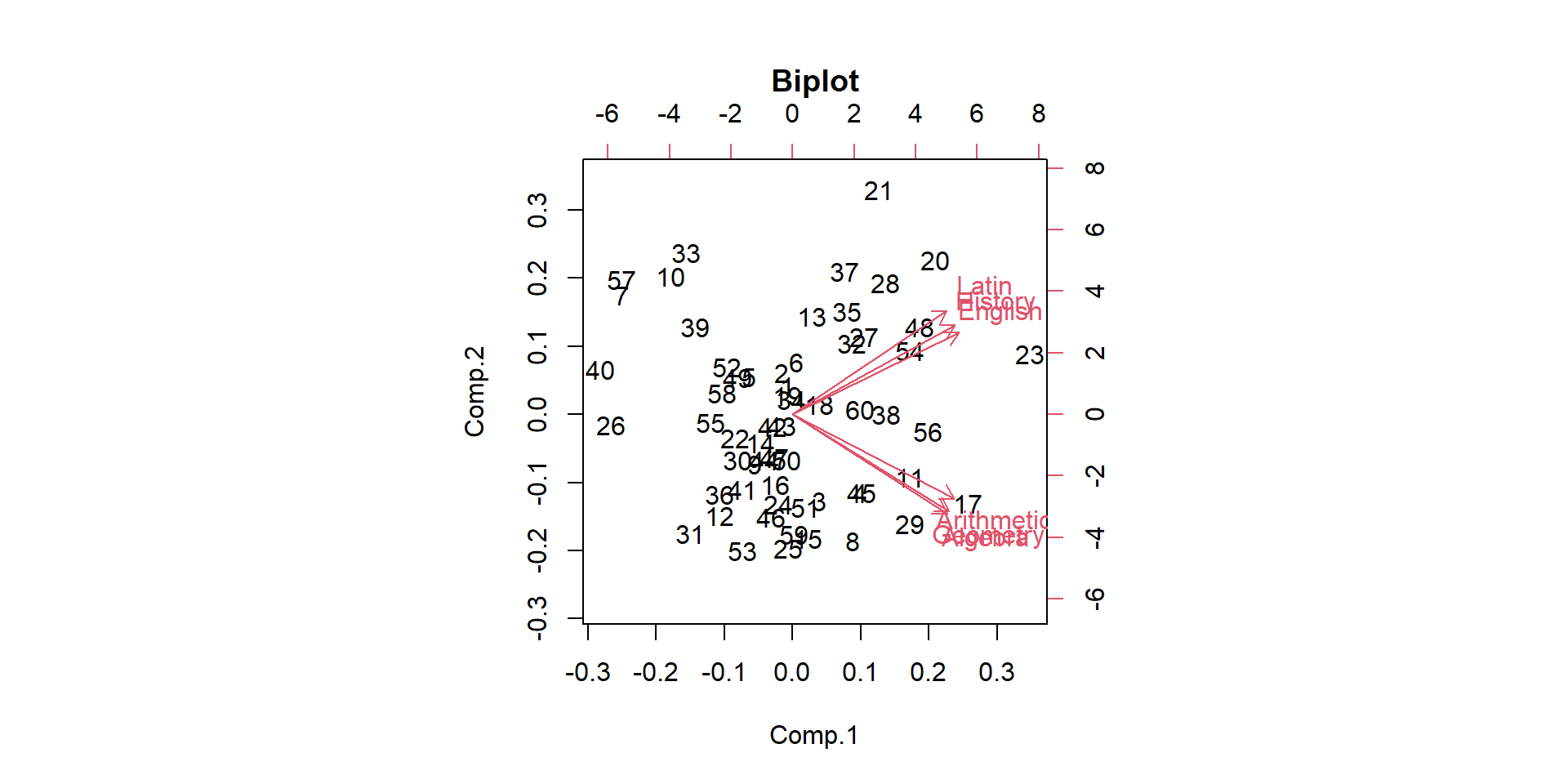
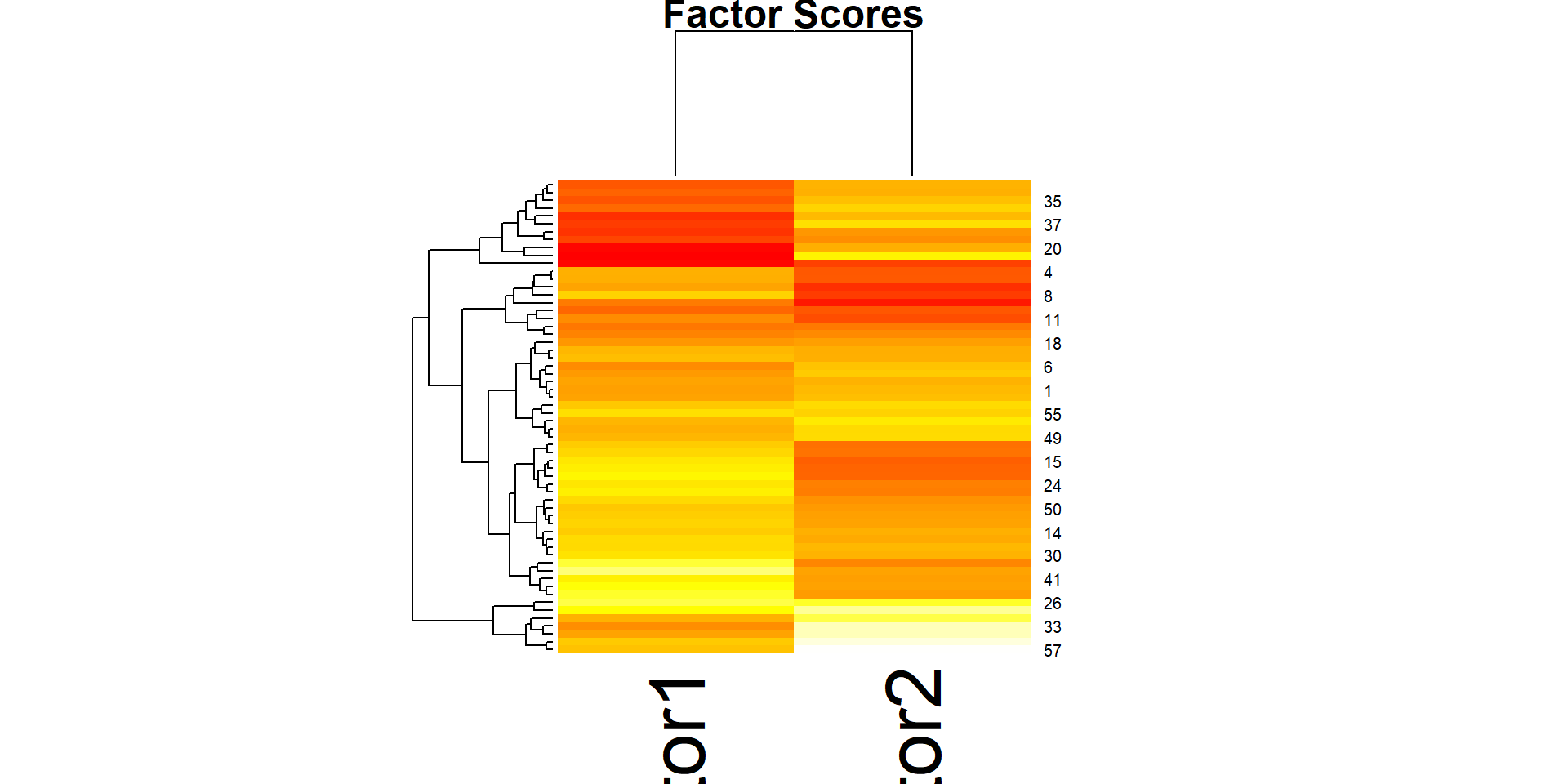
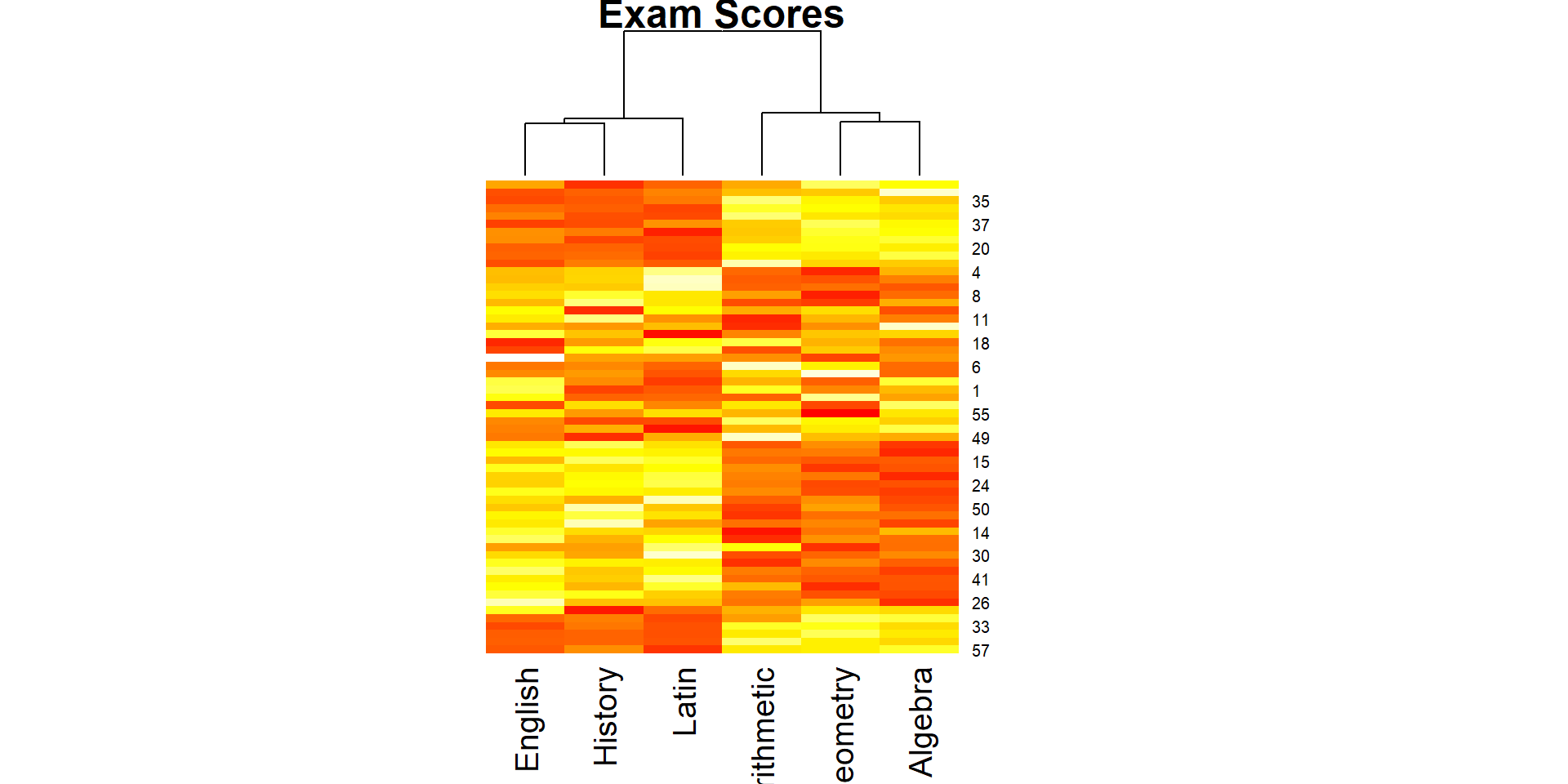
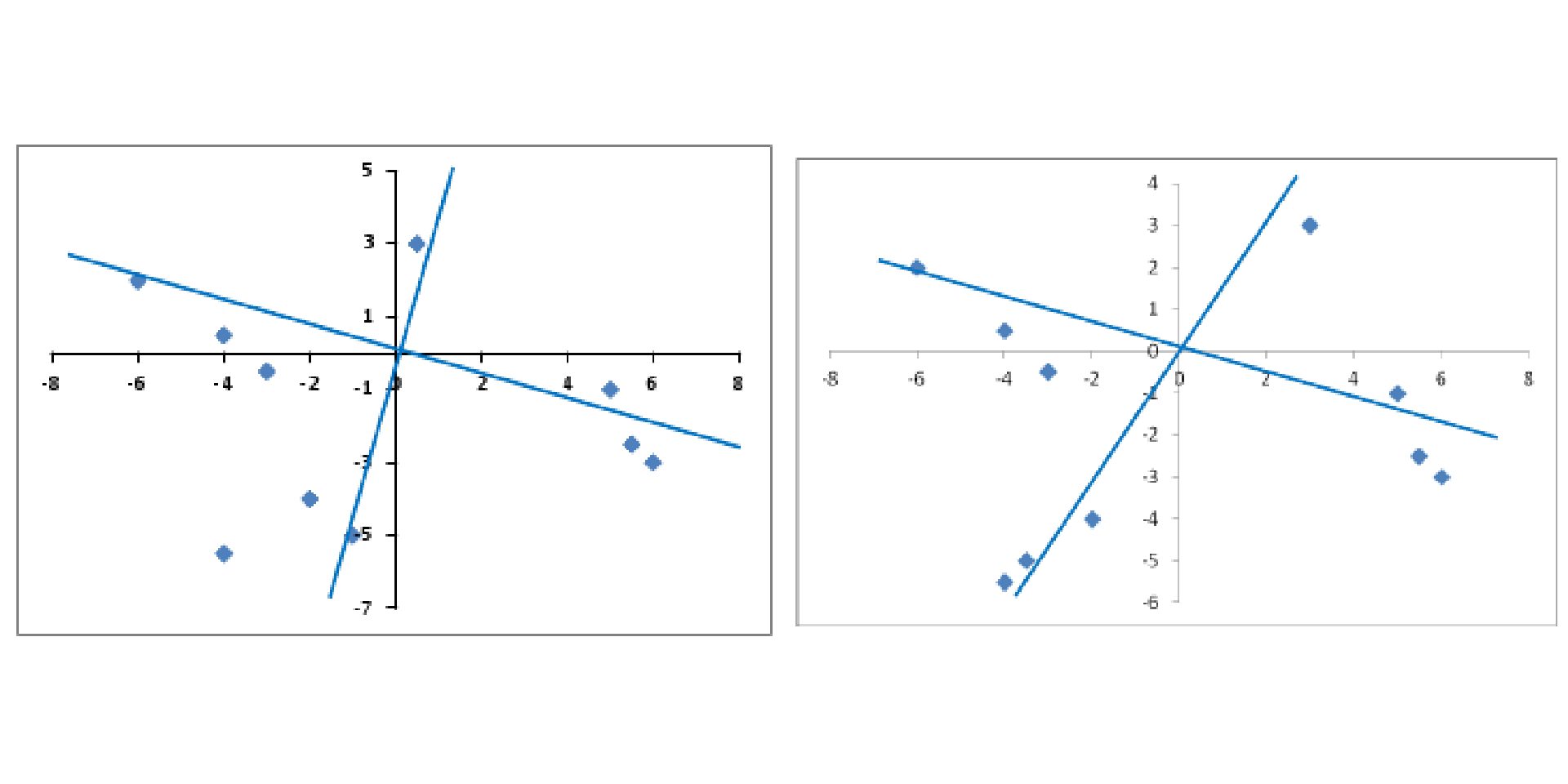
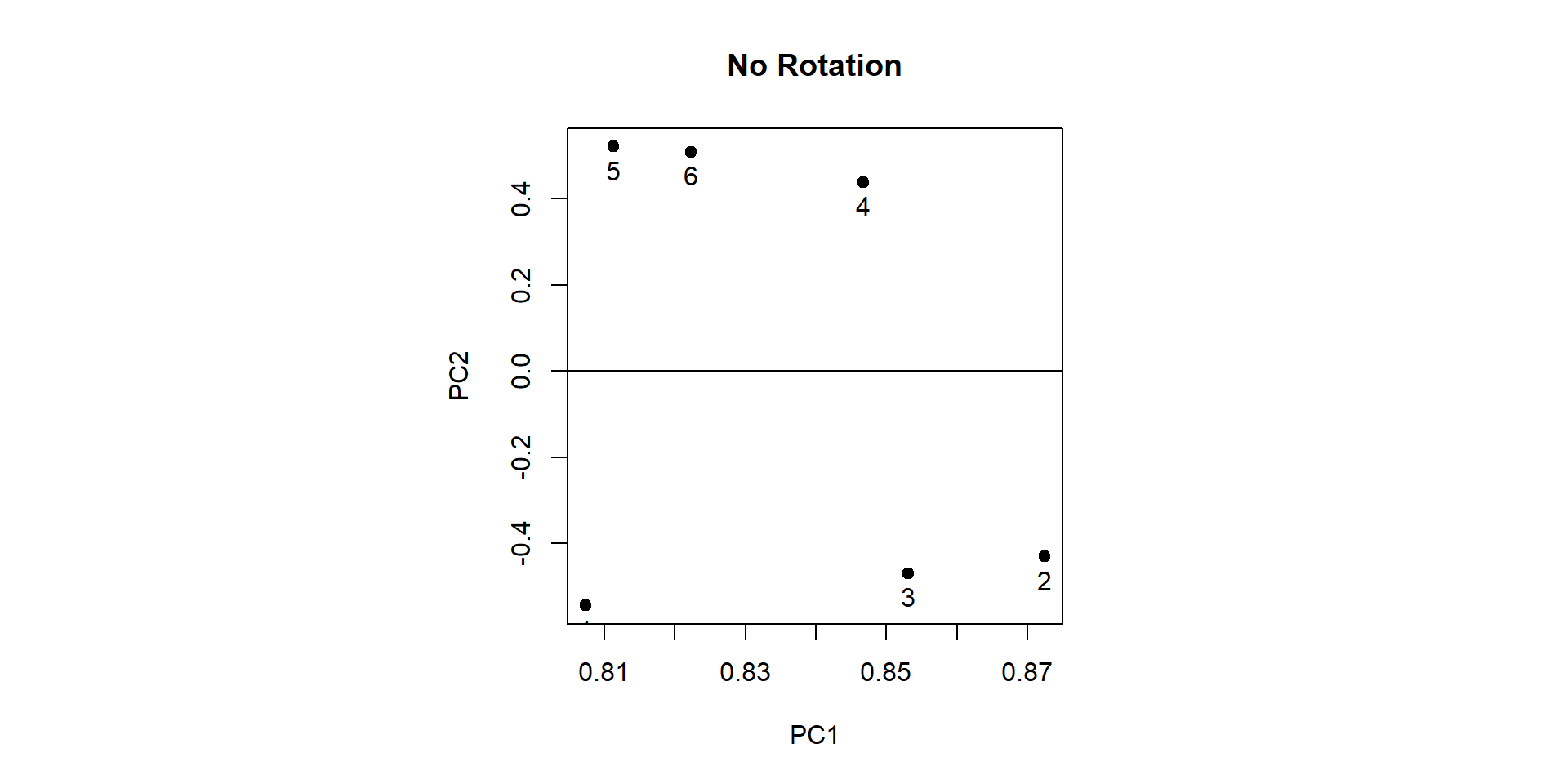
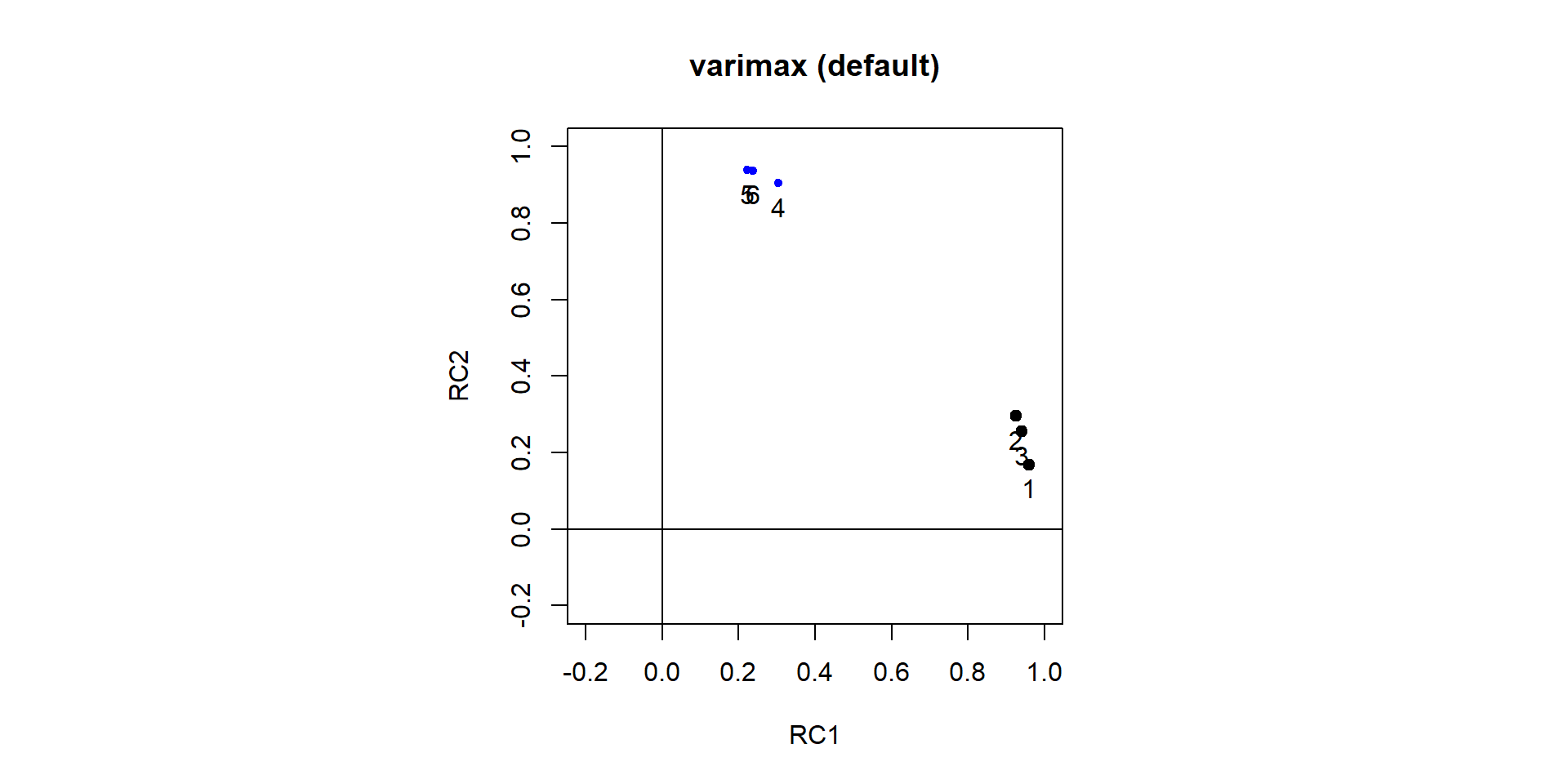
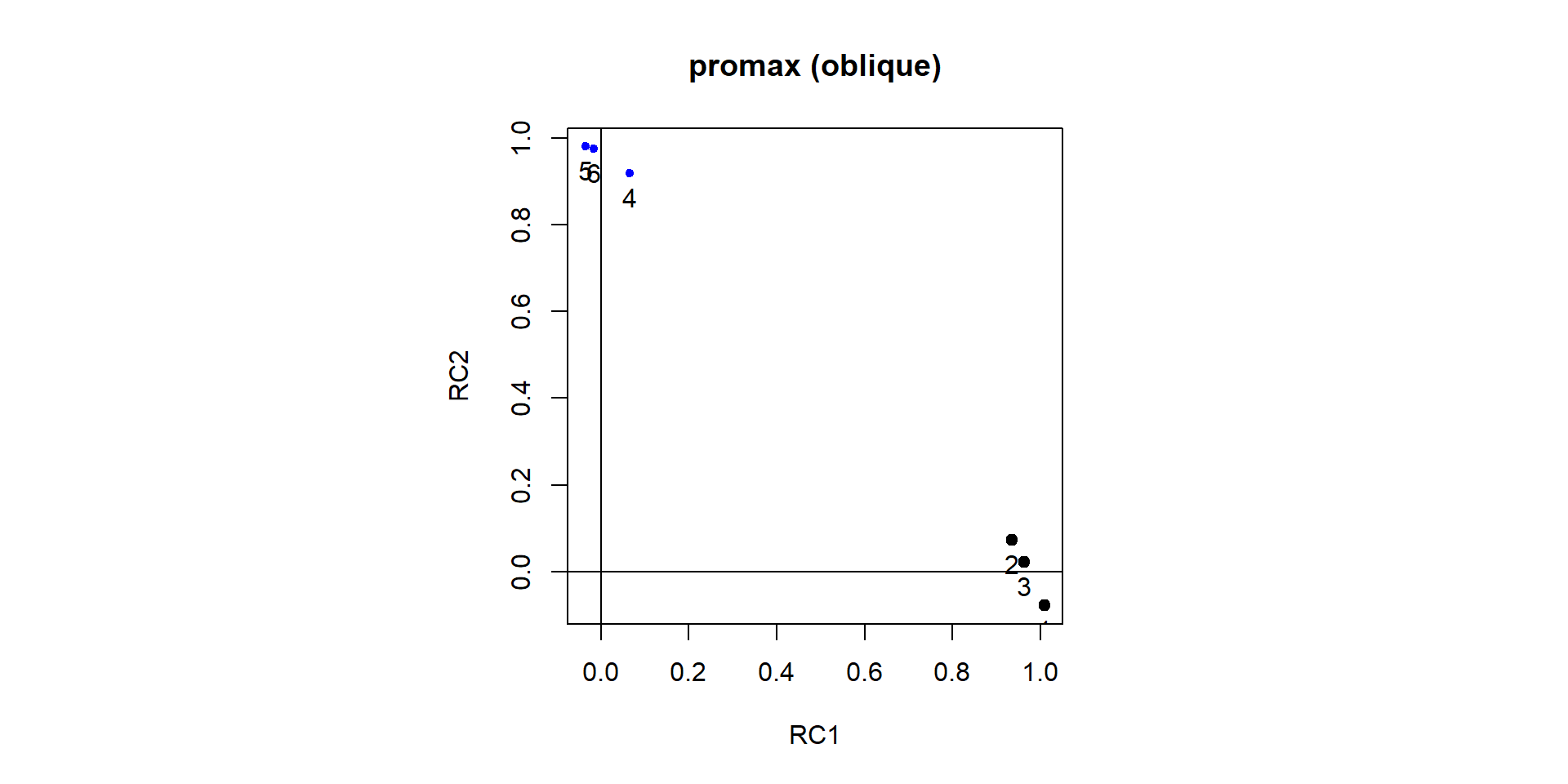
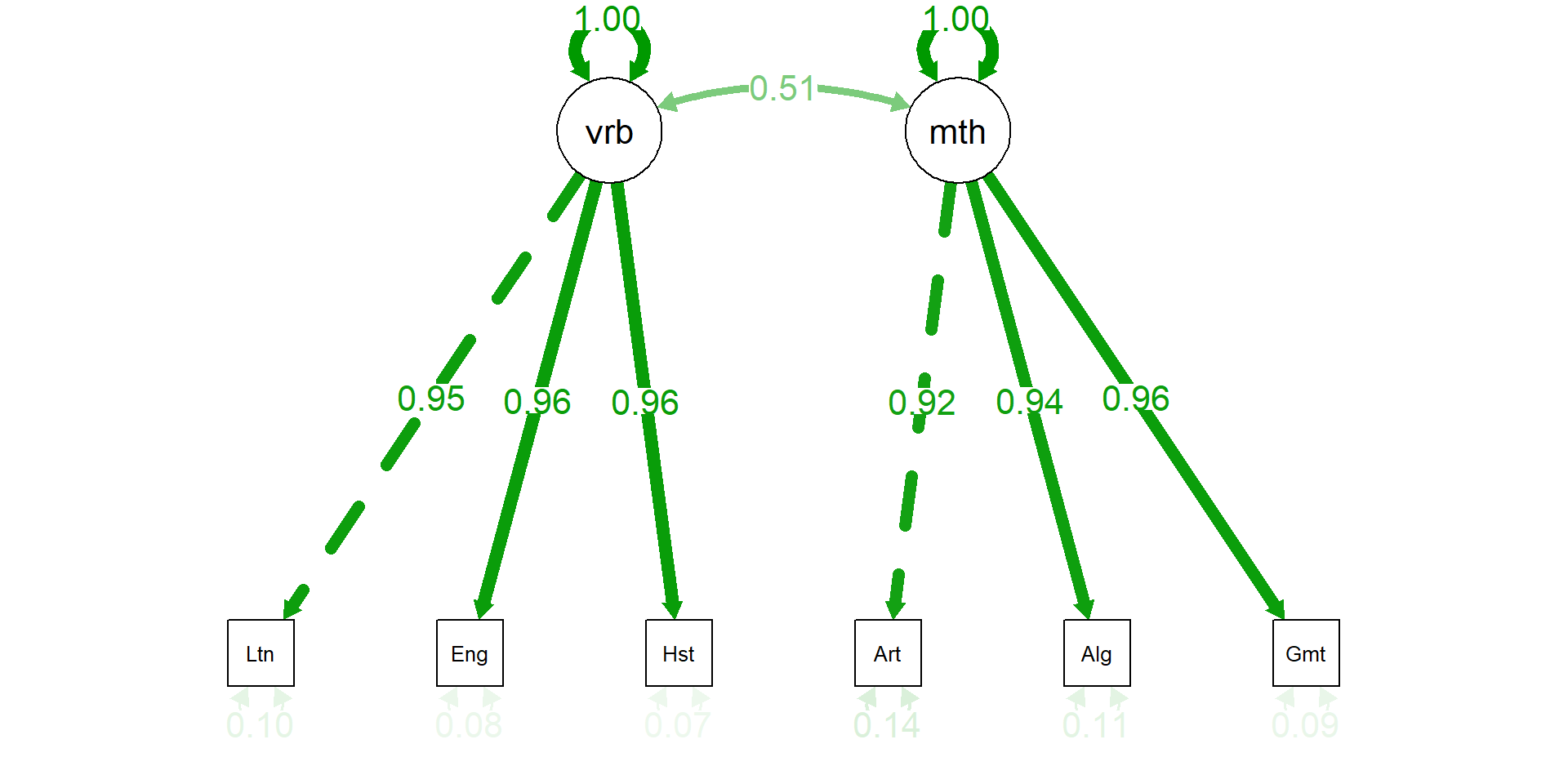
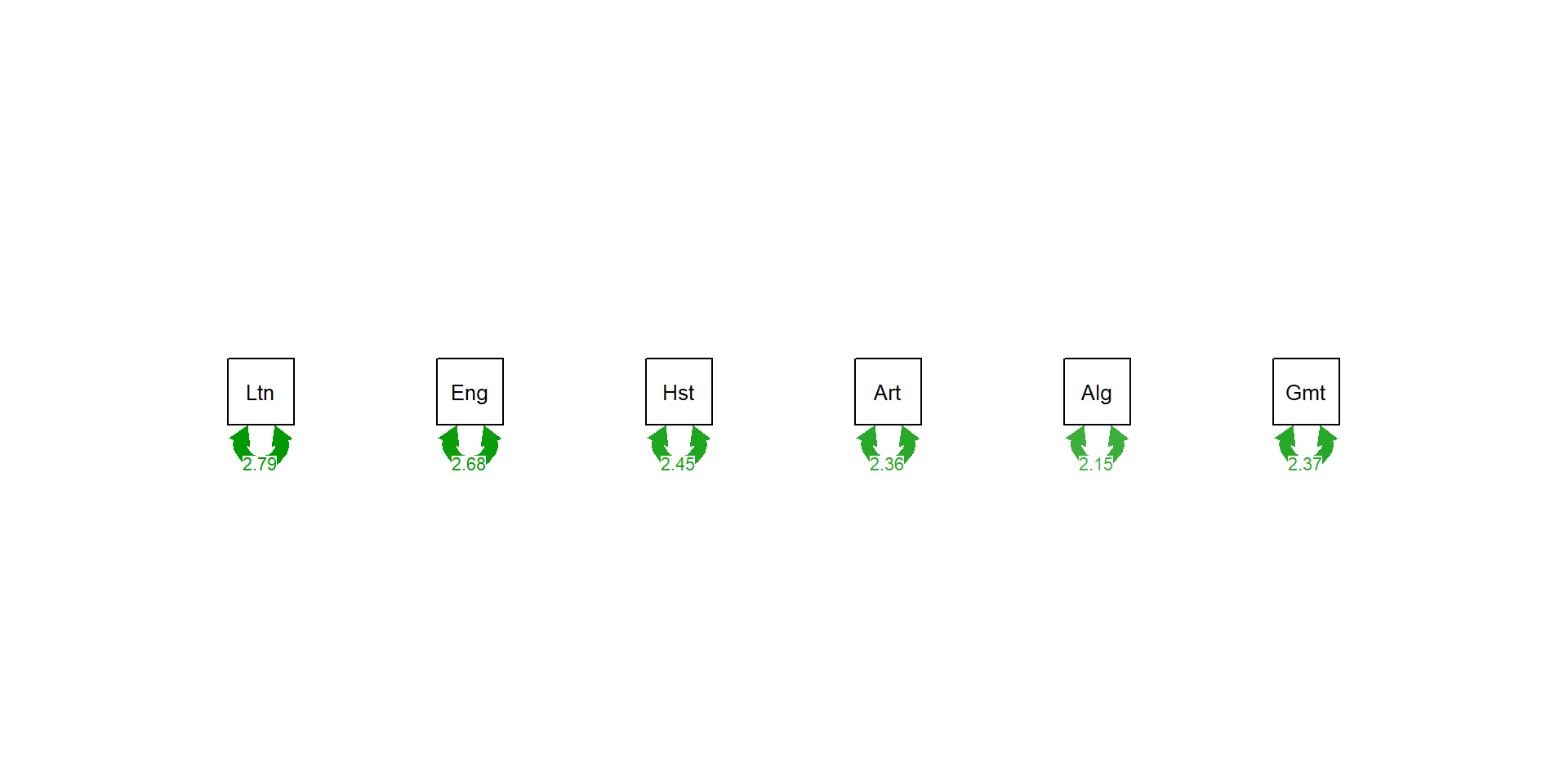
Comp.1 Comp.2
[1,] -0.12510081 0.387885389
[2,] -0.24313883 0.565843287
[3,] 0.61706648 -1.169012826
[4,] 1.57192571 -1.061544977
[5,] -1.01259642 0.511535779
[6,] 0.08159618 0.709633832
[7,] -3.97028171 1.616681077
[8,] 1.40252349 -1.718247894
[9,] -0.87166984 -0.676249351
[10,] -2.83899249 1.873213808
[11,] 2.72771614 -0.850526753
[12,] -1.70338268 -1.375732667
[13,] 0.45223712 1.335051512
[14,] -0.73935793 -0.391321467
[15,] 0.36383804 -1.679191292
[16,] -0.40033457 -0.951024825
[17,] 4.07653952 -1.202123539
[18,] 0.61591640 0.133077527
[19,] -0.12488695 0.259977199
[20,] 3.32163279 2.091207138
[21,] 2.00387753 3.054062570
[22,] -1.33374043 -0.320546882
[23,] 5.52664746 0.816383883
[24,] -0.33635132 -1.217102103
[25,] -0.10498766 -1.814458635
[26,] -4.21673559 -0.146734355
[27,] 1.65883546 1.052902505
[28,] 2.16635789 1.786264476
[29,] 2.73200731 -1.478791682
[30,] -1.27463938 -0.621599441
[31,] -2.37991077 -1.616443270
[32,] 1.37425859 0.969212378
[33,] -2.47024496 2.191557493
[34,] -0.02205375 0.198973539
[35,] 1.27620254 1.394858358
[36,] -1.69777512 -1.082925687
[37,] 1.21645111 1.935638299
[38,] 2.17466798 -0.001283984
[39,] -2.26900283 1.182184153
[40,] -4.47651168 0.610396065
[41,] -1.16212454 -1.013488801
[42,] -0.46563689 -0.170207403
[43,] -0.25281534 -0.155695673
[44,] -0.64299833 -0.617060300
[45,] 1.60383026 -1.065000051
[46,] -0.49803511 -1.390479937
[47,] -0.42374873 -0.582368919
[48,] 2.95636926 1.190798983
[49,] -1.27521832 0.505207025
[50,] -0.14644013 -0.618323482
[51,] 0.29360813 -1.258124936
[52,] -1.53084891 0.644636778
[53,] -1.16723274 -1.853200992
[54,] 2.73613797 0.871310350
[55,] -1.90680059 -0.110926424
[56,] 3.14836985 -0.234451019
[57,] -3.98034871 1.828724734
[58,] -1.63857531 0.287901854
[59,] 0.03921813 -1.629983051
[60,] 1.56468801 0.069052627